
Before using the functionality for 2D structure analysis, the user must select a Consensus Structure to use for the Viewing the 2D structure and Base composition.
Note
This functionality will only work with dot-bracket annotations and will not work if none is given.
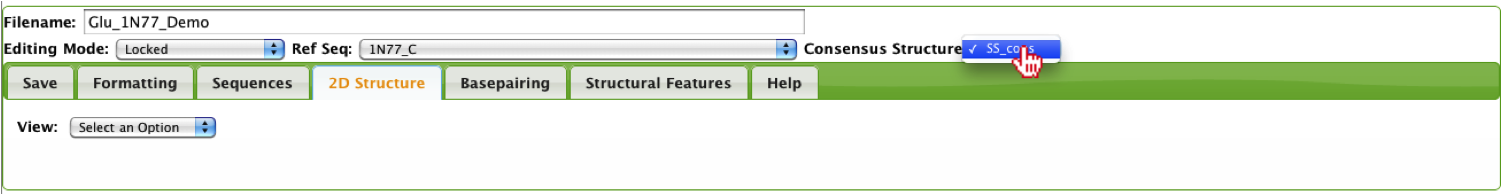
Once the user has selected their reference sequence and selected a sequence from the alignment, they can generate a 2D structure by selecting “2D Structure” from the View menu under the 2D Structure Tab.
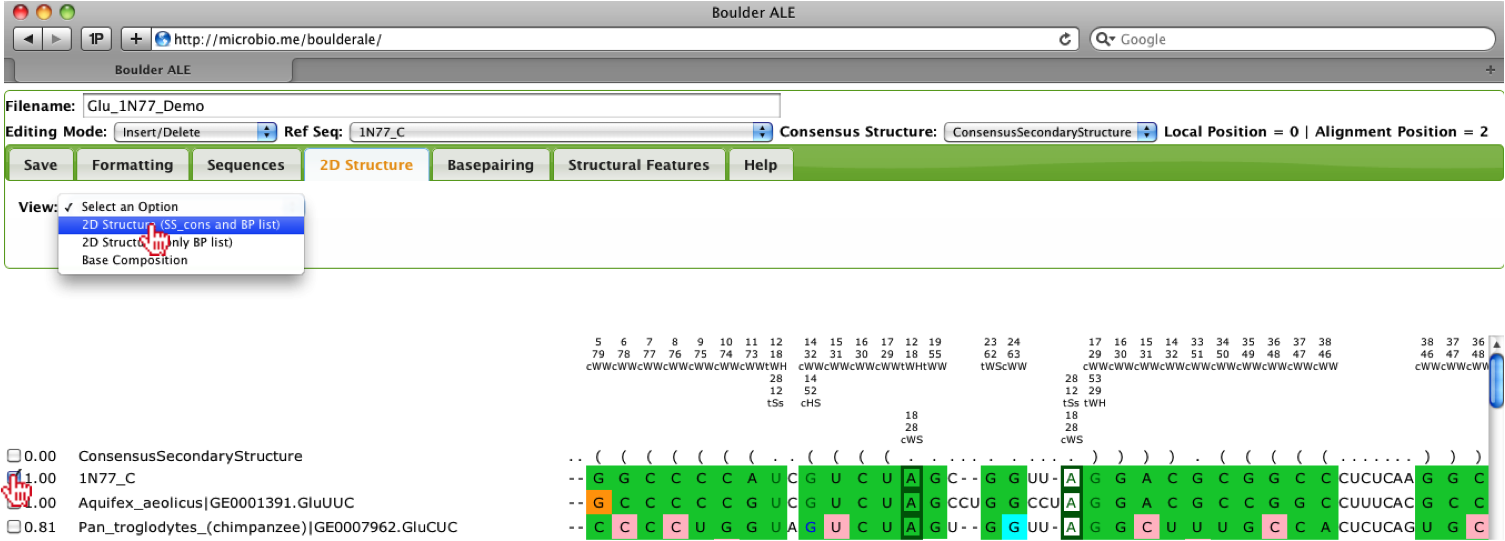
Once the new window appears, the user’s sequence will be displayed in the VARNA java-applet [1].
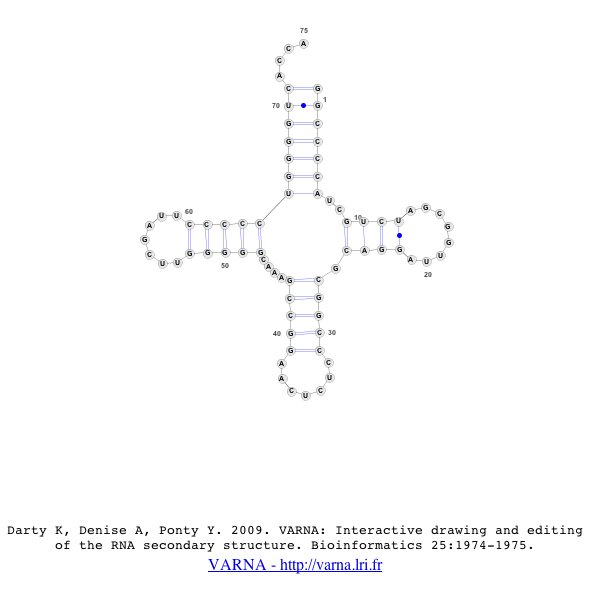
For more information on VARNA, please refer to Darty K, Denise A, Ponty Y, 2009 [1]
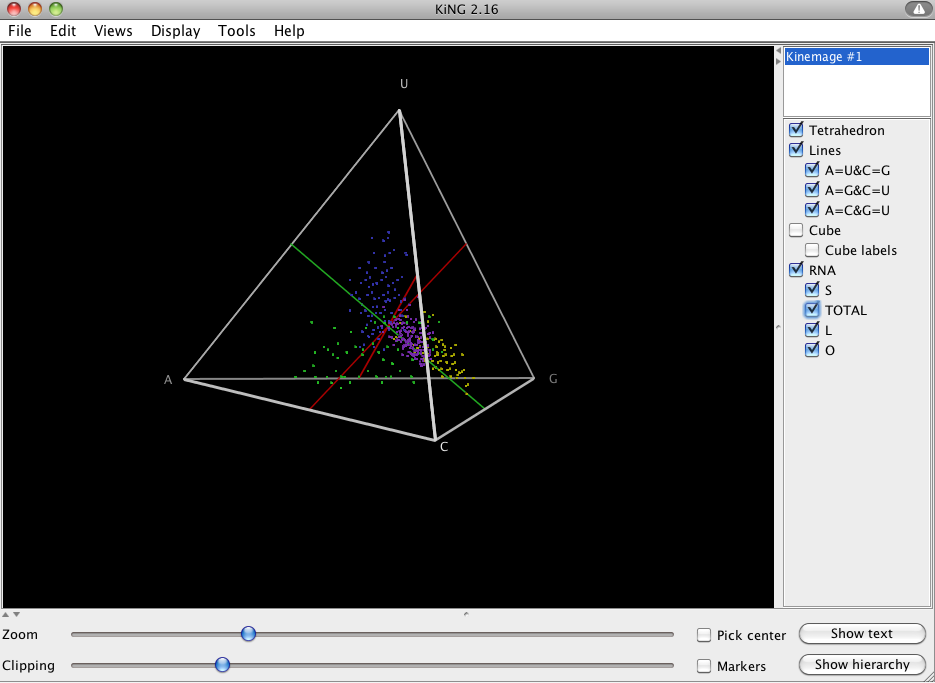
After evaluating the consensus secondary structure in the alignment, the user may be interested in the base composition for stems, loops, or other regions of the alignment [2]. To view the base composition, the user should select “Base Composition” from the View menu under the 2D Structure Tab.
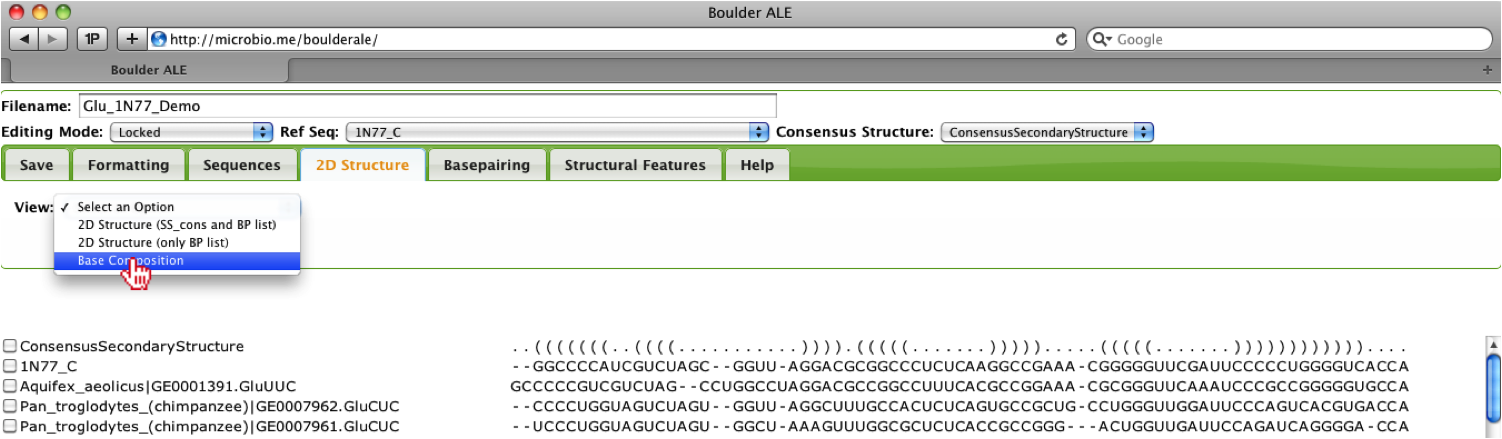
This will generate a kinemage file, which will open in the KiNG java-applet. In the menu you should see “S”, “L”, “O” and “Total” under the RNA menu, where the letters stand for Stem, Loop, Other, and Total, respectively.
For more information on base composition, please refer to Smit S, Knight R, Heringa J, 2009 [2].
| [1] | (1, 2) Darty K, Denise A, Ponty Y. 2009. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics 25:1974-1975. |
| [2] | (1, 2) Smit S, Knight R, Heringa J. 2009. RNA structure prediction from evolutionary patterns of nucleotide composition. Nucleic acids research 37:1378-1386. |