
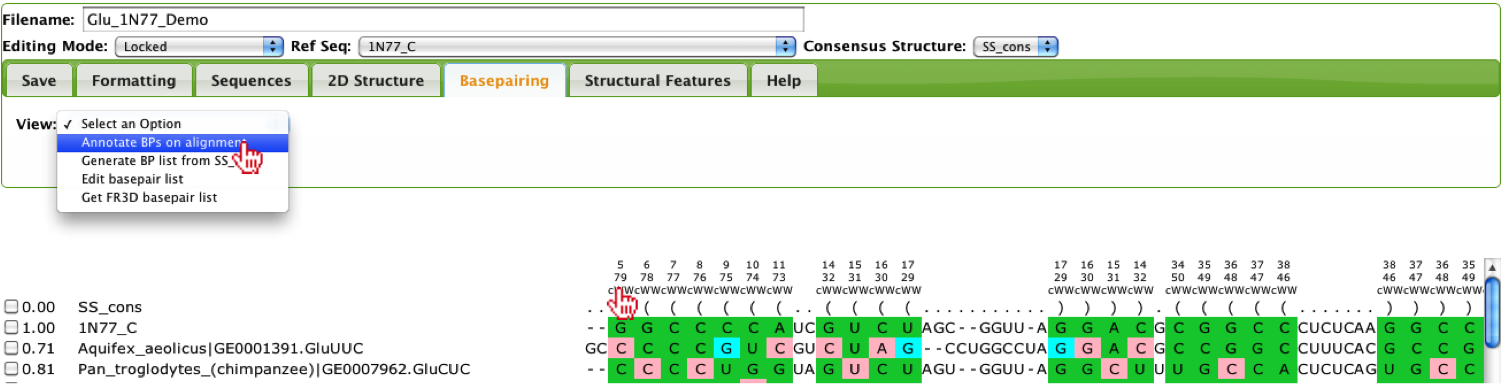
Within the Basepairing tab, the user has the ability to annotate and evaluate their alignment, if a basepair list was given, by selecting the Annotate BPs on Alignment option in the select-box. If no basepair list was given, the user can either download a basepair list if a PDB structure is known by select the Get FR3D basepair list option or they can generate a basepair list given a consensus secondary structure, by selecting the Generate BP list from SS_cons.
If FR3D basepair list is given, the alignment will be annotated with Watson-Crick and non-Watson-Crick basepairs using the Leontis-Westhof nomenclature [1], which are listed above the alignment. Since each base can make up to 3 basepair interactions, so there are three rows of basepairs, where the first row lists the first basepair that base is involved in, and the second row and third rows show additional basepairs the base is involved in. If basepair list is based on the consensus secondary structure, only the Watson-Crick interactions will be listed. The coloring of the cells correspond to the row the annotation appears in above the alignment, where the background of the cell is colored to correspond to the first row of basepair annotations, the text of the cell is colored when a basepair is listed in the second row of basepair annotations and the border of the cell corresponds to basepair annotations in the third row. The coloring corresponds whether the base participates in a basepair interaction that is isosteric (green), non-isosteric (pink) or not allowed (cyan) when compared to the Reference Sequence. Isostericity is based on the IsoDiscrepancy Index proposed by Stombaugh et al., 2009 [2].
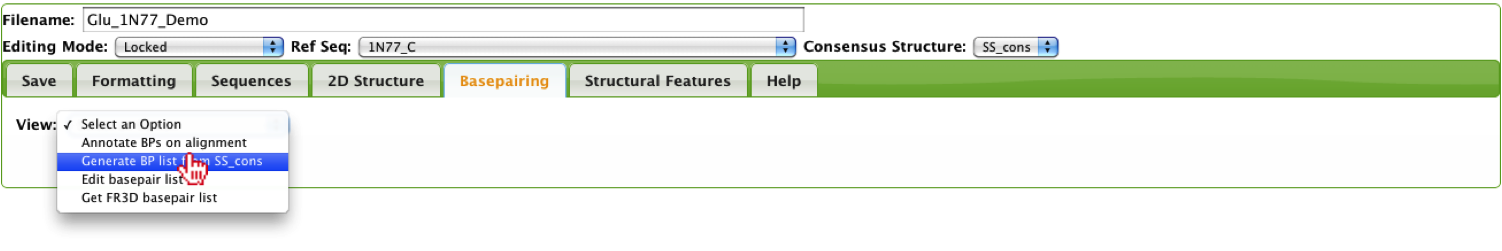
This option will parse the user-defined consensus structure and will create a list of basepairs based on the bracket annotation.
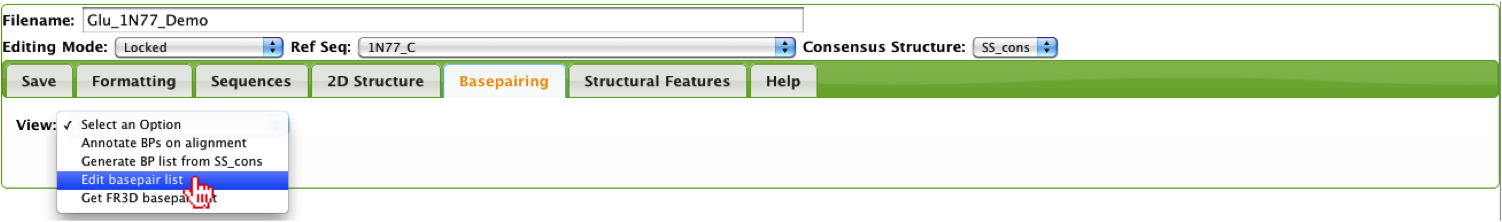
This option allows the user to manually edit the basepair list, where they can add/remove basepairs. When defining a basepair list, the reference sequence is required and cannot be changed without generating a new basepair list. The reason is that basepair coloring is based on whether a basepair is isosteric to the reference.
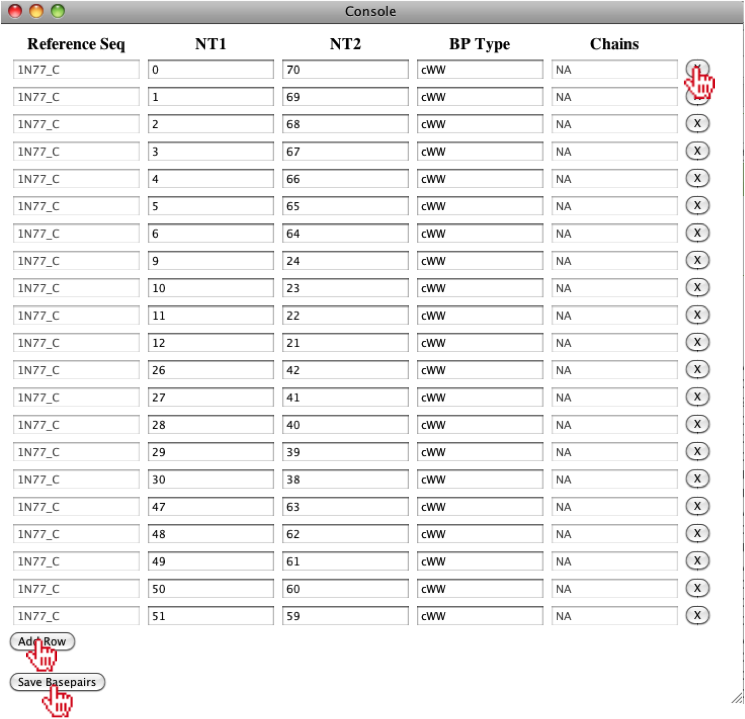
Once the new window is opened, the user can either edit the basepairs directly, or remove/add basepairs using the buttons to the right/bottom of the window.
If the user’s alignment has a corresponding PDB structure, the user can get the FR3D basepair list and sequence from the PDB.
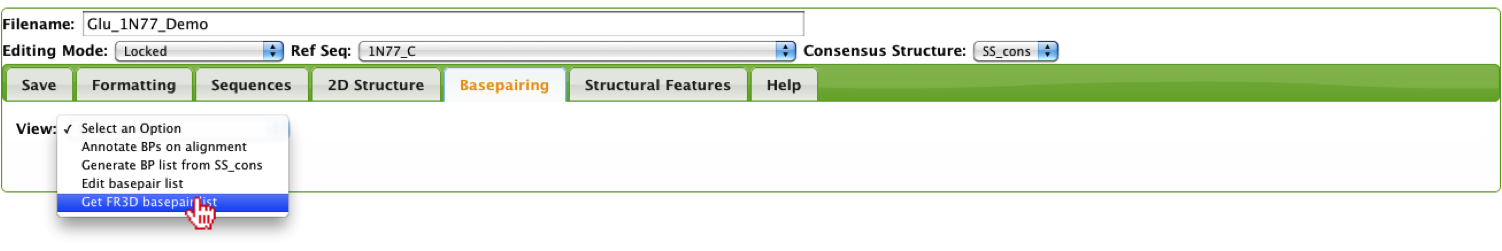
This option allows the user to enter a PDB and it returns a basepair list along with the sequence for each chain.
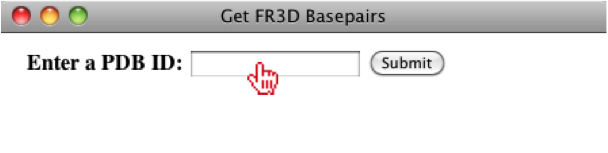
Once the user has selected the chain(s) they would like to use for their basepair list, they can optionally add the sequence if it is not already in the alignment.
Note
If more than one chain is interacting, then the user will be given multiple options for concatenating the two sequences, however; if only one chain interacts with itself, only one sequence will be listed.
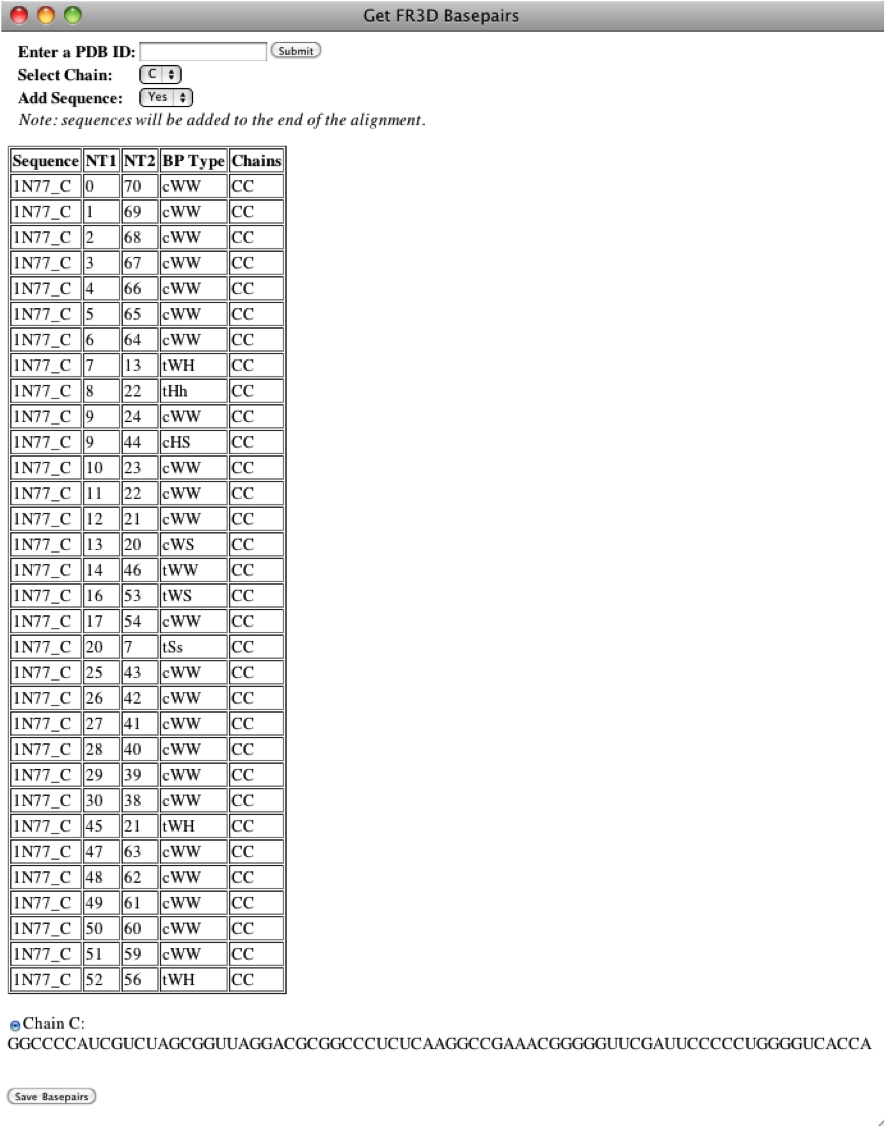
Note
The reference sequence must be in the alignment to evaluate isostericity, so most likely you will need to add the sequence. If the user appends the PDB sequence, it will be added to the end of the alignment.
| [1] | Leontis, N. B., & Westhof, E. (2001). Geometric nomenclature and classification of RNA base pairs. RNA, 7(4), 499-512. |
| [2] | Stombaugh, J., Zirbel, C. L., Westhof, E., & Leontis, N. B. (2009). Frequency and isostericity of RNA base pairs. Nucleic Acids Res, 37(7), 2294-2312. |